Product Info
This kit is an enhanced version of EZ-editor™ Monoclone Genotype Validation Kit (extraction free), which is specially designed to solve the problem of difficult validation of long or complex DNA sequences. It has th e advantages of microscale, high efficiency and accuracy . The minimum sample amount is about 30-50 cells , which allow the high-throughput validation of cell samples on 96 -well plates. The genomic DNA can be released by directly lysing cell sample s, so the preparation of the cell genome sample takes as soon as 15 minutes. It can be used in the PCR experiment without DNA extraction and purif ication, which is easy to operate and more efficient in high-throughput operation!
In addition, the specially developed PCR reagent CloneAmp LA Mix (+Dye) is not only highly compatible with the factors that may affect the PCR reaction, such as the culture medium residue and cell lysis components in the genome sample, but also has a very high amplification efficiency. Using the cell lysis solution treated with MicroCell DNA Lysis as the template, a 10 kb DNA fragment can be amplified within 1.5 hours, and the accurate validation of the genotype of the monoclonal cells can be effectively achieved.
After a lot of tests and optimization, this kit is applicable to all animal-derived cells, including adherent cells, suspended cells, human cells, mouse cells, rat cells, etc. The recommended sample size is 3x103~105 cells (generally, one well of a 96-well plate contains 1x104 cells), and the minimum sample size is not less than 30-50 cells. In addition, Ubigene provides an exclusive design tool for identifying primers for gene editing target sites (EZ-editor™ primer design platform) and a tool for interpreting sequencing results of gene editing samples (EZ-editor™ genotype analysis system). Massive experimental data are automatically processed and analyzed, allowing you to easily identify the genotype of gene editing samples.
Kit Components
Components | YK-MVH-50 | YK-MVH-100 | YK-MVH-250 | Storage temperature |
MicroCell DNA Lysis | Buffer HpA | 5 mL | 10 mL | 25 mL | Normal |
Buffer HpB | 0.5 mL | 1 mL | 2.5 mL | Normal |
PCR Validation | CloneAmp LA Mix(+Dye) | 625 μL | 1.25 mL | 3.125 mL | -20℃ |
CloneAmp Ctrl | 100 μL | 200 μL | 500 μL | -20℃ |
GC Buffer | 250 μL | 500 μL | 1.25 mL | -20℃ |
Example
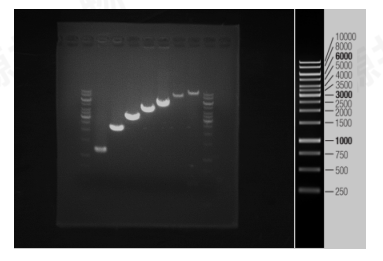
Figure 1. This image shows the results of PCR amplification of templates with different lengths, taking 5 μL PCR product to electrophoresis, the size of the target bands are 1200 bp, 2000 bp, 3000 bp, 4000 bp, 5000 bp, 8000 bp, 10000 bp respectively. The amplified bands in the gel are bright and clear, and the amplification efficiency is excellent. The genomic DNA can be directly used as the PCR template without purification, and the maximum amplification band is 10000 bp.







